

Using GraphSAGE embeddings for downstream classification model
source link: https://towardsdatascience.com/using-graphsage-embeddings-for-downstream-classification-model-4492e01ae54e
Go to the source link to view the article. You can view the picture content, updated content and better typesetting reading experience. If the link is broken, please click the button below to view the snapshot at that time.
Using GraphSAGE embeddings for downstream classification model
Learn how to use the GraphSAGE embeddings in Neo4j Graph Data Science library to improve your Machine Learning workflows
The use of knowledge graphs and graph analytics pipeline is getting more and more popular. If you keep an eye on the graph analytics field, you already know that graph neural networks are trending. Unfortunately, there aren’t many tutorials out there on how to use them in a practical application. For this reason, I have decided to write this blog post, where you will learn how to train a convolutional graph neural network and integrate it into your machine learning workflow to improve downstream classification model accuracy.
Agenda
In this example, you will reproduce the protein role classification task from the original GraphSAGE article. The task is to classify protein roles in terms of their cellular function across various protein-protein interaction graphs (PPI). The dataset contains 22 PPI graphs, with each graph corresponding to a different human tissue. The average PPI graph contains 2373 nodes, with an average degree of 28.8. There are available predefined positional gene sets, motif gene sets, and immunological signatures for each protein in the network. Based on those features and their connections, you will predict the roles of proteins in the network. You will train both the classification and GraphSAGE model on 20 graphs and then average prediction F1 scores on two test graphs.
Graph model
As mentioned, we are dealing with a protein-protein interaction network. This is a monopartite network, where nodes represent proteins and relationships represent their interactions.

Additionally, the protein nodes have the predefined features stored as a property. The embeddings_all property contains all 50 features stored as a list of floats. I have also prepared the decoupled properties, where the embedding_x property holds a single feature and x ranges from 0 to 49. You will see later in the blog post why the decoupled properties are required. The protein nodes also contain a secondary label that could be either Train or Test. With the help of the secondary label, you can easily perform the train-test data split.
Neo4j environment setup
To set up the Neo4j environment, you will first need to download and install the Neo4j Desktop application. You don’t need to create a database instance just yet. To avoid bugging you with the import process, I have prepared a database dump file, which you can go ahead and load a database instance from it. In this example, I used the Neo4j 4.2.0 version. If you aren’t familiar with how to load a Neo4j database instance from a dump file, you can take a look at my blog post for more detailed instructions. Next, you will need to install both the APOC and the Graph Data Science libraries. Using the Neo4j Desktop application, you can install both libraries with a single click as shown below.

Now that you have loaded the database instance and installed both libraries, you can go ahead and start the database. Run the following query in the Neo4j Browser interface to make sure the graph is loaded correctly.
MATCH p=(:Protein)-[:INTERACTS]->(:Protein)
RETURN p LIMIT 25
Results

You should see a similar visualization in your Neo4j Browser. The green nodes represent the proteins and the relationships represent the interactions between the proteins. You can also notice that a protein can interact with itself. That is represented with self-loops, where the relationship starts and points to the same node.
Classification using predefined features
To get a baseline f1 score, you will first train the classification model using only the predefined features available for proteins. The code is identical to the code found in the official GraphSAGE repository, where they used the Stochastic Gradient Descent classifier model to train and predict protein roles. The only difference is that here you will be fetching the data from a Neo4j database instance.
The baseline f1 score, where you used only the predefined features of proteins, is 0.422. Let’s now try to improve the classification model accuracy using the GraphSAGE algorithm
GraphSAGE algorithm
GraphSAGE is a convolutional graph neural network algorithm. The key idea behind the algorithm is that we learn a function that generates node embeddings by sampling and aggregating feature information from a node’s local neighborhood. As the GraphSAGE algorithm learns a function that can induce the embedding of a node, it can also be used to induce embeddings of a new node that wasn’t observed during the training phase. This is called inductive learning.

If you want to learn more about the training process and the math behind the GraphSAGE algorithm, I suggest you take a look at the An Intuitive Explanation of GraphSAGE blog post by Rıza Özçelik or the official GraphSAGE site.
Using GraphSAGE embeddings for a downstream classification task
Neo4j Graph Data Science library operates entirely on heap memory to enable fast caching for the graph’s topology, containing only relevant nodes, relationships, and weights. Graph algorithms are executed on an in-memory projected graph model, which is separate from Neo4j’s stored graph model.
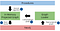
Before you can execute any graph algorithms, you have to project the in-memory graph via the Graph Loader component. You can use either native projection or cypher projection to load the in-memory graph.
In this example, you will use the native projection feature to load the in-memory graph. To start, you will project the training data and store it as a named graph in the Graph Catalog. The current implementation of the GraphSAGE algorithm supports only node features that are of type Float. For this reason, you will include the decoupled node properties ranging from embedding_0 to embedding_49 in the graph projection instead of a single property embeddings_all, which holds all the node features in the form of a list of Floats. Additionally, you will treat the projected graph as undirected.
UNWIND range(0,49) as i
WITH collect('embedding_' + toString(i)) as embeddingsCALL gds.graph.create('train','Train',
{INTERACTS:{orientation:'UNDIRECTED'}},
{nodeProperties:embeddings})
YIELD graphName, nodeCount, relationshipCount
RETURN graphName, nodeCount, relationshipCount
Next, you will train the GraphSAGE model. The model’s hyper-parameter settings were mostly copied from the original paper. I have noticed that the lower learning-rate setting had the most impact on the downstream classification accuracy. Another import hyper-parameter is the samplingSizes parameter, where the size of the list determines the number of layers (defined as K parameter in the paper), and the values determine how many nodes will be sampled by the layers. Find more information about the available hyper-parameters in the documentation.
UNWIND range(0,49) as i
WITH collect('embedding_' + toString(i)) as embeddingsCALL gds.beta.graphSage.train('train',{
modelName:'proteinModel',
aggregator:'pool',
batchSize:512,
activationFunction:'relu',
epochs:10,
sampleSizes:[25,10],
learningRate:0.0000001,
embeddingDimension:256,
featureProperties:embeddings})
YIELD modelInfo
RETURN modelInfo
The training process took around 20 minutes on my laptop. After the training process finishes, the GraphSAGE model will be stored in the model catalog. You can now use this model to induce node embeddings on any projected graph with the same node properties used during the training. Before testing the downstream classification accuracy, you have to load the test data as an in-memory graph in the Graph Catalog.
UNWIND range(0,49) as i
WITH collect('embedding_' + toString(i)) as embeddingsCALL gds.graph.create('test','Test',
{INTERACTS:{orientation:'UNDIRECTED'}},
{nodeProperties:embeddings})
YIELD graphName, nodeCount, relationshipCount
RETURN graphName, nodeCount, relationshipCount
With the GraphSAGE model trained and both the training and test data projected as an in-memory graph, you can go ahead and calculate the f1 score using the GraphSAGE embeddings in a downstream classification model. Remember, the GraphSAGE model has not observed the test data during the training phase.
Using the GraphSAGE embeddings as feature input to the classification model, you have improved the f1 score to 0.462. You can also try to follow the other examples in the original GraphSAGE paper to hone your graph data science skills.
Takeaways
- Connections within your data can help you increase the accuracy of your ML models
- GraphSAGE algorithm can induce embeddings of new unseen nodes, without the need for re-training process
As always, the code is available on GitHub.
References
Recommend
About Joyk
Aggregate valuable and interesting links.
Joyk means Joy of geeK