

Predicting Hospital Readmission with Deep Learning from Scratch and with Keras
source link: https://towardsdatascience.com/predicting-hospital-readmission-with-deep-learning-from-scratch-and-with-keras-309efc0f75fc?gi=5af8888d4ee3
Go to the source link to view the article. You can view the picture content, updated content and better typesetting reading experience. If the link is broken, please click the button below to view the snapshot at that time.
Let’s use deep learning to identify patients at risk for readmission!

Nov 24 ·9min read

Introduction
Recently, I watched a video https://www.youtube.com/watch?v=JPBz7-UCqRo ) from my friend Eric Ma on the fundamentals of deep learning. To teach deep learning, he breaks it into 3 key ingredients: model, loss function, and optimization routine. Throughout the tutorial, he uses an automatic differentiation toolbox. However, I find it deeply satisfying to do the derivatives myself (at least for simple cases). Today, I thought I would build a 2-layer neural network from scratch following Eric’s approach but with algebraic derivatives (from Andrew Ng’s Coursera class ), and then implement it again using Keras (a deep learning framework).
Data Set
For this project, we will use the same dataset as my previous post on prediction hospital admission with the diabetes hospital dataset from UCI ( https://archive.ics.uci.edu/ml/datasets/diabetes+130-us+hospitals+for+years+1999-2008 ). For review of this project and the feature engineering, see my prior post https://towardsdatascience.com/predicting-hospital-readmission-for-patients-with-diabetes-using-scikit-learn-a2e359b15f0
Project Definition
Predict if a patient with diabetes will be readmitted to the hospital within 30 days.
Feature Engineering
We will start this post as if we already completed the feature engineering section of my previous post which includes creating numerical, categorical (one-hot encoding), and ordinal features. These features are conveniently saved with the prior notebook and are included in my github repo .

For deep learning, it is important to fill in missing values and normalize the data. We will use SimpleImputer and StandardScaler to do this from scikit-learn.

From Scratch
Here we would like to build a simple two-layer neural network from scratch using the structure introduced by Eric Ma:
- Model
- Loss Function
- Optimization Routine
I will try to stick to the notation introduced by Andrew Ng in his Coursera specialization ( https://www.coursera.org/specializations/deep-learning ).
Model
The model we will use is a two-layer neural network as shown below:

Here we will have n_x input variables, n_1 hidden nodes, and an output node including m samples. For this model we will use logistic regression for the activation function in the hidden layer nodes as well as in the output layer.

Here our activation functions will have the following form

We will use a vectorized notation for more efficient calculation. In this notation, the first column of X will be all the features for the first sample (note this is opposite of what is loaded in Python at this point, so we will need to Transpose the X matrices).
The parameters we will use for this model will have the following dimensions
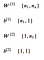
For simplicity, let’s choose n1 = 64 nodes to approximately halve the number of input variables. Following Eric’s notation, let’s save all these parameters in a dictionary. We will initialize these randomly since setting to 0 will not work as the weights would all be the same node to node.
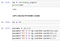
To calculate our estimate of y_hat for our m
examples, we can feed the information forward through the model with the following equations (note the dimensions are in curly braces).

We can write this in code using the following feedforward function. Here we will leave the activation function for the hidden layer as an variable to the function.

Loss function
Now that we have a method for calculating y_hat given some parameters, we need to find the ‘best’ parameters. In order to define ‘best’, we need a cost function to define how good the parameters are. The loss function we use for binary classification is:

Super obvious where this comes from right?
I prefer to see where this comes from, so let’s take a short detour and derive this equation. If we think about the output of our model as a probability of y given x, we can write the following for a single example:

Which can be written more cleverly as:
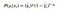
If we assume all of our samples are independent, the likelihood of seeing all of our data is the product of the individual probabilities:

Now all we have to do is find the parameters that maximizes this likelihood. That sounds quite complicated given this has the product term. Fortunately, maximizing the log of the likelihood function also maximizes the likelihood function (since log is monotonically increasing). Before we do this, let’s remind ourselves about a property of logs:

Applying to our likelihood function gives us:

This is close to our cost function! The only difference is we multiply by a -1 and divide by m (the number of samples). The negative multiplication switches it from maximization to minimization problem.

For our optimization routines, we will need the derivative of this cost function J
. Eric did this with the python package jax.grad
dlogistic_loss = grad(logistic_loss)
but I would like to write this out explicitly to better understand the math.
To do this derivative, we actually work from right to left in our neural network, a process known as Backpropagation. We can do this with derivative fundamentals: CHAIN RULE!
Before we dive into this, let’s pull some function derivatives we will use
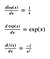
and linearity
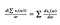
Since our cost sums over each sample, for now let’s just drop the notation for each sample from the calculations (and the 1/m multiplier). Here we will use short-hand notation from Andrew Ng with definitions :=
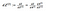
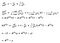
Now we can take derivatives for the parameters in the output layer (and properly taking into account the matrix math):
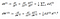
To move backward in the network and considering any activation function g^[layer](Z[layer])
. Here *
represents element-wise multiplication and comes into play because we are using chain rule for a single sample.

As you can see here, there is a clear pattern that would allow us to expand this to any number of hidden layers.
Let’s write our back propagation function now that takes as inputs the parameters, a function for derivative of activation function, the feedforward values, and the output values.

Here d_logistic
is the derivative of the logistic function with respect to z
.
At this point it is good to verify that our parameters and d_params have the same shapes for each parameter set.
Optimization Routine
We will use gradient descent to update our parameters. Gradient descent basically works by updating the parameters iteratively by moving the opposite direction of the gradient by the learning amount alpha
. We can run this in a for loop and keep track of the losses. Note tqdmn allows us to see the progress with a progress bar (kind of neat, thanks Eric!).
We can verify the losses decrease over iterations:

We can then calculate our predictions for the training and validation sets:

Using scikit-learn metrics, we can plot the ROC curve:

The training of this by-scratch model was quite slow. Let’s use Keras instead which has additional more efficient optimization routines like Adam. Keras is also great for building more complex networks in just a few lines of code.
Keras
First let’s import some packages

We will need to adjust our output labels just a bit for Keras to have one column for each label:

Now we can build our model using Sequential. Here I will use the ReLu activation function instead of logistic function since ReLu tends to work better. I will also add in Dropout which is a form of regularization that helps reduce overfitting.

Here the final output layer has two nodes (one for each label). These outputs get normalized with the softmax function to convert scores into probabilities.
We then compile the model with the loss function and optimizer specified. Here ‘categorical_crossentropy’ is the formula for our loss function defined above. This also can be expanded to include any number of outcomes

Now we fit the model with

There are two input parameters here batch_size
and epochs
that are included with the fitting. Batch size indicates how many samples are used in each iteration. In our by-scratch implementation we included all samples in each iterations, which was took more time to compute. If you run the calculation with a smaller batch, you are able to iterate more quickly. Epoch is defined as how many times you iterate over the entire dataset after breaking it into smaller batches.
Similar to scikit-learn, we get the predictions with predict proba and just grab the second column:

I played around with different numbers of nodes in the hidden layer, drop rates and additional layers. The final model then has a validation AUC = 0.66 as shown in the ROC below:

Unfortunately, this has the same performance as all the other models we trained in the prior post!
Conclusion
In this post we trained a 2 layer neural network from scratch and with Keras. Please comment below if you have any questions. The code is on my github repo: https://github.com/andrewwlong/diabetes_readmission_deep
Recommend
About Joyk
Aggregate valuable and interesting links.
Joyk means Joy of geeK