

Hyperparameter tuning for Random Forests
source link: https://andrewpwheeler.com/2022/10/10/hyperparameter-tuning-for-random-forests/
Go to the source link to view the article. You can view the picture content, updated content and better typesetting reading experience. If the link is broken, please click the button below to view the snapshot at that time.
Hyperparameter tuning for Random Forests
Motivated to write this post based on a few different examples at work. One, we have periodically tried different auto machine learning (automl) libraries at work (with quite mediocre success). They are OK for a baseline, not so much for production. Two, a fellow data scientist was trying some simple hyperparameter search in random forests, and was searching over all the wrong things.
For those not familiar, automl libraries, such as data robot or databricks automl, typically do a grid search over different models given a particular problem (e.g. random forest, logistic regression, XGBoost, etc.). And within this you can have variants of similar models, e.g. RFMod1(max-depth=5) vs RFMod2(max-depth=10). Then given a train/test split, see which model wins in the test set, and then suggests promoting that model into production.
My experience at work with these different libraries (mostly tabular medical records data), they choose XGBoost variants at a very high frequency. I think this is partially due to poor defaults for the hyperparameter search in several of these automl libraries. Here I will just be focused on random forests, and to follow along I have code posted on Github.
Here I am using the NIJ recidivism challenge data I have written about in the past, and for just some upfront work I am loading in various libraries/functions (I have the recid functions in the githup repo). And then making it easy to just load in the train/test data (with some smart feature engineering already completed).
from datetime import datetime
import matplotlib.pyplot as plt
import numpy as np
import optuna
import pandas as pd
from sklearn.metrics import brier_score_loss, roc_auc_score
from sklearn.model_selection import train_test_split
from sklearn.ensemble import RandomForestClassifier
import recid # my custom NIJ recid funcs
# Read in data and feature engineering
train, test = recid.fe()Now first, in terms of hyperparameter tuning you need to understand the nature of the model you are fitting, and how those hyperparameters interact with the model. In terms of random forests for example, I see several automl libraries (and my colleague) doing a search over the number of trees (or estimators in sklearn parlance). Random forests the trees are independent, and so having more trees is always better. It is sort of like saying would you rather me do 100 simulations or 1,000,000 simulations to estimate a parameter – the 1,000,000 will of course have a more accurate estimate (although may be wasteful, as the extra precision is not needed).
So here, using the NIJ defined train/test split, and a set of different fixed parameters (close to what I typically default to for random forests). I show how AUC or the Brier Score changes with the number of trees, over a grid from 10 to 1000 trees:
# Loop over tree sizes
ntrees = np.arange(10,1010,10)
auc, bs = [], []
# This is showing number of estimators is asymptotic
# should approach best value with higher numbers
# but has some variance in out of sample test
for n in ntrees:
print(f'Fitting Model for ntrees:{n} @ {datetime.now()}')
# Baseline model
mod = RandomForestClassifier(n_estimators=n, max_depth=5, min_samples_split=50)
# Fit Model
mod.fit(train[recid.xvars], train[recid.y1])
# Evaluate AUC/BrierScore out of sample
pred_out = mod.predict_proba(test[recid.xvars])[:,1]
auc.append(roc_auc_score(test[recid.y1], pred_out))
bs.append(brier_score_loss(test[recid.y1], pred_out))
# Making a plot of the
fig, (ax1, ax2) = plt.subplots(2, figsize=(6,8))
ax1.plot(ntrees, auc)
ax1.set_title('AUC')
ax2.plot(ntrees, bs)
ax2.set_title('Brier Score')
plt.savefig('Ntree_grid.png', dpi=500, bbox_inches='tight')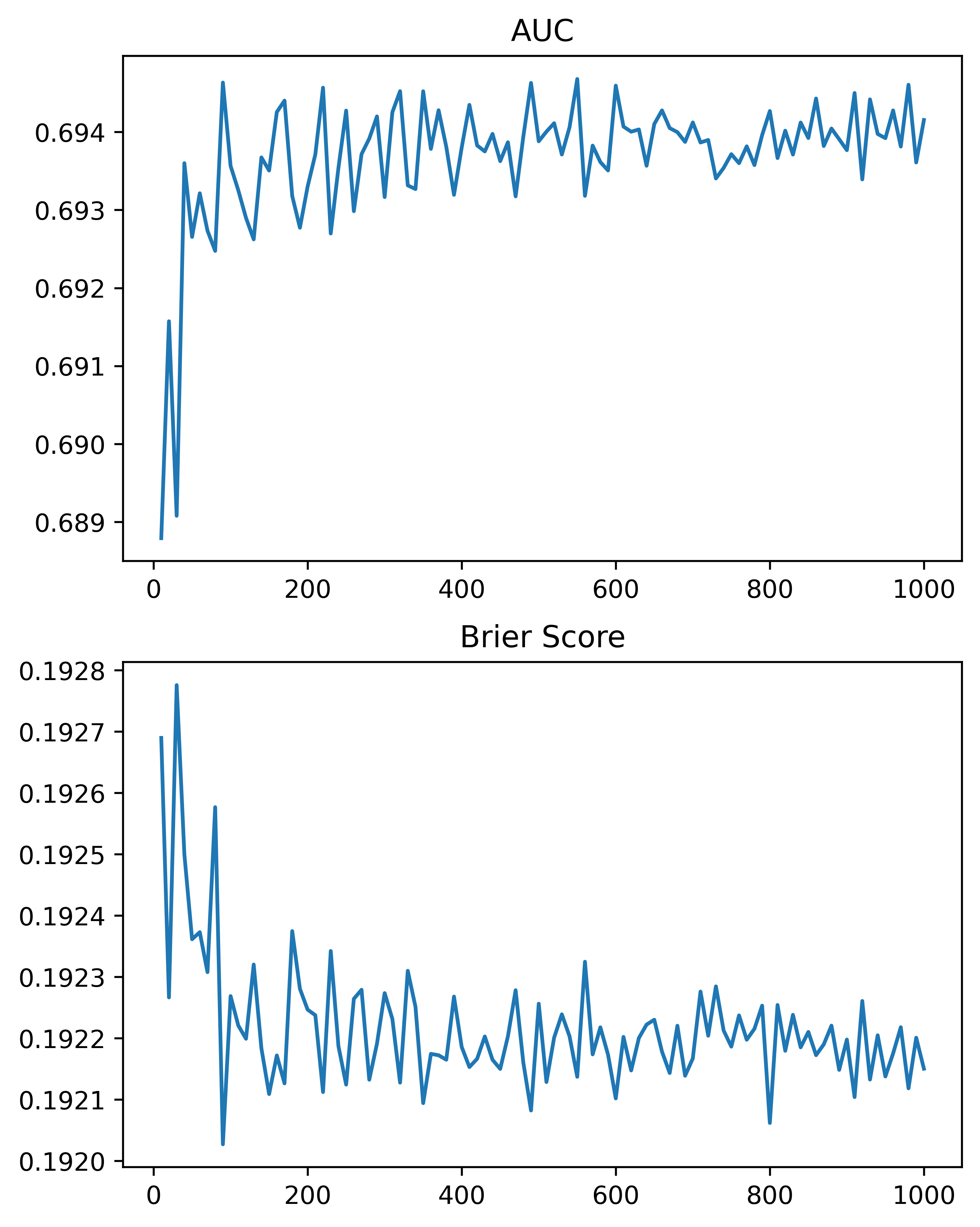
We can see that the relationship is noisy, but the trend clearly decreases with tree size, and perhaps asymptotes post 200 some trees for both metrics in this particular set of data.
So of course it depends on the dataset, but when I see automl libraries choosing trees in the range of 10, 50, 100 for random forests I roll my eyes a bit. You always get more accurate (in a statistical sense), with more trees. You would only choose that few for convenience in fitting and time savings. The R library ranger has a better default of 500 trees IMO (sklearns 100 is often too small in doing tests). But there isn’t much point in trying to hyperparameter tune this – your hyperparameter library may choose a smaller number of trees in a particular run, but this is due to noise in the tuning process itself.
So what metrics do matter for random forests? All machine learning models you need to worry about over-fitting/under-fitting. This depends on the nature of the data (number of rows, can fit more parameters, fewer columns less of opportunity to overfit). Random forests this is dependent on the complexity of the trees – more complex trees can overfit the data. If you have more rows of data, you can fit more complex trees. So typically I am doing searches over (in sklearn parlance) max-depth (how deep a tree can grow), min-samples-split (can grow no more trees is samples are too tiny in a leaf node). Another parameter to search for is how many columns to subsample as well (more columns can find more complex trees).
It can potentially be a balancing act – if you have more samples per leaf, it by default will create less complex trees. Many of the other hyperparameters limiting the complexity of the trees are redundant with these as well (so you could really swap out with max-depth). Here is an example of using the Optuna library a friend recommended to figure out the best parameters for this particular data set, using a train/eval approach (so this splits up the training set even further):
# Consistent train/test split for all evaluations
tr1, tr2 = train_test_split(train, test_size=2000)
def objective(trial):
param = {
"n_estimators": 500,
"max_depth": trial.suggest_int("max_depth", 2, 10),
"min_samples_split": trial.suggest_int("min_samples_split", 10, 200, step=10),
"max_features": trial.suggest_int("max_features", 3, 15),
}
mod = RandomForestClassifier(**param)
mod.fit(tr1[recid.xvars], tr1[recid.y1])
pred_out = mod.predict_proba(tr2[recid.xvars])[:,1]
auc = roc_auc_score(tr2[recid.y1], pred_out)
return auc
study = optuna.create_study(direction="maximize")
study.optimize(objective, n_trials=100)
trial = study.best_trial
print(f"Best AUC {trial.value}")
print("Best Params")
print(trial.params)So here it chooses fairly deep trees, 9, but limited complexity via the sample size in a leaf (90). The number of features per tree is alsio slightly larger than default (here 25 features, so default is sqrt(25)). So it appears my personal defaults were slightly under-fitting the data.
My experience regression prefers smaller depth of trees but lower sample sizes (splitting to 1 is OK), but for binary classification limiting sample sizes in trees is preferable.
One thing to pay attention to in these automl libraries, a few do not retrain on the full dataset with the selected hyperparameters. For certain scenarios you don’t want to do this (e.g. if using the eval set to do calibrations/prediction intervals), but if you don’t care about those then you typically want to retrain on the full set. Another random personal observation, random forests really only start to outperform simpler regression strategies at 20k cases, with smaller sample sizes and further splitting train/eval/test, the hyperparameter search is very noisy and mostly a waste of time. If you only have a few hundred cases just fit a reasonable regression model and call it a day.
So here I retrain on the full test set, and then look at the AUC/Brier Score compared to my default model.
# Lets refit according to these params for the full
# training set
mod2 = RandomForestClassifier(n_estimators=500,**trial.params)
mod2.fit(train[recid.xvars], train[recid.y1])
pred_out = mod2.predict_proba(test[recid.xvars])[:,1]
auc_tuned = roc_auc_score(test[recid.y1], pred_out)
bs_tuned = brier_score_loss(test[recid.y1], pred_out)
print(f"AUC tuned {auc_tuned:.4f} vs AUC default {auc[-1]:.4f}")
print(f"Brier Score tuned {bs_tuned:.4f} vs default {bs[-1]:.4f}")It is definately worth hyperparameter tuning once you have your feature set down, but lift of ~0.01 AUC (which is typical for hypertuned tabular models in my experience) is not a big deal with initial model building. Figuring out a smart way to encode some relevant feature (based on domain knowledge of the problem you are dealing with) typically has more lift than this in my experience.
Recommend
About Joyk
Aggregate valuable and interesting links.
Joyk means Joy of geeK