
1

MOEAD原理及Python实现、MOEAD实现、基于分解的多目标进化、 切比雪夫方法-(python完...
source link: https://www.cnblogs.com/Twobox/p/16416509.html
Go to the source link to view the article. You can view the picture content, updated content and better typesetting reading experience. If the link is broken, please click the button below to view the snapshot at that time.
MOEAD原理及Python实现、MOEAD实现、基于分解的多目标进化、 切比雪夫方法-(python完整代码)
确定某点附近的点
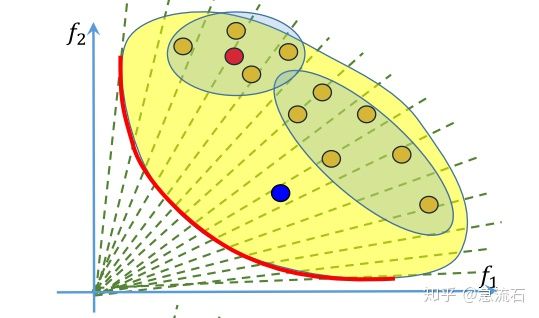
答:每个解对应的是一组权重,即子问题,红点附近的四个点,也就是它的邻居怎么确定呢?由权重来确定,算法初始化阶段就确定了每个权重对应的邻居,也就是每个子问题的邻居子问题。权重的邻居通过欧式距离来判断。取最近的几个。
取均匀分布向量
https://www.cnblogs.com/Twobox/p/16408751.html
MOEAD实现
算法理解与流程
https://www.zhihu.com/question/263555181?sort=created
其中两个回答都挺好的
1. 输入N m
# N表示取点密度 m表示问题维度
1.1 输入 T
# 表示取最近的T个作为邻居
2. 生成均匀分布权重向量
2.1 计算每个权重向量之间的欧拉距离
3. 权重向量个数即为:初始种群个数
4. 初始化种群,每个个体一一对应权重
4.1 更具权重之间距离,取前T个作为邻居person
5. EP = 空
# 维护成最优前沿
6. 计算最初的全局最优Z
# 把每个带入f1 f2中,取最小值 z1 z2
7. 开始循环N代
7.1对于每个个体,在领域中选取2个个体进行交叉变异,获得2个新个体
7.1.1更新全局解z
7.2在领域中随机选择2个个体,用新个与旧个体进行对比
# 新个体带入子目标问题,直接对比值即可
7.3如果更优,则替换旧个体dna
7.4更新EP
# 如果有别接收的新解,将新解与EP每一个进行比较,删除被新解支配的,如果新解没有被旧解支配,那么加入EP
代码实现设计
# 分析
需要维护的数据结构:
某个体最近的T位邻居: 可以考虑采用对象列表即可
均匀分布的权重向量:一个二维ndarray数组即可
权重向量与个体对应关系:个体对象,直接保存权重向量数组
权重向量之间的距离矩阵:开局初始化,不变的
EP list,里面的个体是对象的引用
z list
目标函数集合,F list domain list
# 接口设计
class Person
attribute:
dns:一维ndarray
weight_vector: 一维ndarray
neighbor: list<Person>
o_func:Objective_Function 目标函数
function:
mutation
cross_get_two_new_dna:返回2段新dna
compare#与子代比较
accept_new_dna
choice_two_person:p1,p2
class Moead_Util
attribute:
N
M
T:
o_func:Objective_Function
pm:变异概率
EP:[dna1,dna2,..]
weight_vectors:二维数组
Euler_distance:二维数组
pip_size
Z:[] # 这里面元素为一维ndarray数组,即dna,即解
function:
init_mean_vector:二维数组
init_Euler_distance:二维数组
init_population:[]
init_Z:一维属猪
update_ep
update_Z
class Objective_Function:
attribute:
F:[]
domain:[[0,1],[],[]]
function:
get_one_function:Objective_Function
Person.py
1 import numpy as np 2 3 4 class Person: 5 def __init__(self, dna): 6 self.dna = dna 7 self.weight_vector = None 8 self.neighbor = None 9 self.o_func = None # 目标函数 10 11 self.dns_len = len(dna) 12 13 def set_info(self, weight_vector, neighbor, o_func): 14 self.weight_vector = weight_vector 15 self.neighbor = neighbor 16 self.o_func = o_func# 目标函数 17 18 def mutation_dna(self, one_dna): 19 i = np.random.randint(0, self.dns_len) 20 low = self.o_func.domain[i][0] 21 high = self.o_func.domain[i][1] 22 new_v = np.random.rand() * (high - low) + low 23 one_dna[i] = new_v 24 return one_dna 25 26 def mutation(self): 27 i = np.random.randint(0, self.dns_len) 28 low = self.o_func.domain[i][0] 29 high = self.o_func.domain[i][1] 30 new_v = np.random.rand() * (high - low) + low 31 self.dna[i] = new_v 32 33 @staticmethod 34 def cross_get_two_new_dna(p1, p2): 35 # 单点交叉 36 cut_i = np.random.randint(1, p1.dns_len - 1) 37 dna1 = p1.dna.copy() 38 dna2 = p2.dna.copy() 39 temp = dna1[cut_i:].copy() 40 dna1[cut_i:] = dna2[cut_i:] 41 dna2[cut_i:] = temp 42 return dna1, dna2 43 44 def compare(self, son_dna): 45 F = self.o_func.f_funcs 46 f_x_son_dna = [] 47 f_x_self = [] 48 for f in F: 49 f_x_son_dna.append(f(son_dna)) 50 f_x_self.append(f(self.dna)) 51 fit_son_dna = np.array(f_x_son_dna) * self.weight_vector 52 fit_self = np.array(f_x_self) * self.weight_vector 53 return fit_son_dna.sum() - fit_self.sum() 54 55 def accept_new_dna(self, new_dna): 56 self.dna = new_dna 57 58 def choice_two_person(self): 59 neighbor = self.neighbor 60 neighbor_len = len(neighbor) 61 idx = np.random.randint(0, neighbor_len, size=2) 62 p1 = self.neighbor[idx[0]] 63 p2 = self.neighbor[idx[1]] 64 return p1, p2
Objective_Function
1 from collections import defaultdict
2
3 import numpy as np
4
5
6 def zdt4_f1(x_list):
7 return x_list[0]
8
9
10 def zdt4_gx(x_list):
11 sum = 1 + 10 * (10 - 1)
12 for i in range(1, 10):
13 sum += x_list[i] ** 2 - 10 * np.cos(4 * np.pi * x_list[i])
14 return sum
15
16
17 def zdt4_f2(x_list):
18 gx_ans = zdt4_gx(x_list)
19 if x_list[0] < 0:
20 print("????: x_list[0] < 0:", x_list[0])
21 if gx_ans < 0:
22 print("gx_ans < 0", gx_ans)
23 if (x_list[0] / gx_ans) <= 0:
24 print("x_list[0] / gx_ans<0:", x_list[0] / gx_ans)
25
26 ans = 1 - np.sqrt(x_list[0] / gx_ans)
27 return ans
28
29 def zdt3_f1(x):
30 return x[0]
31
32
33 def zdt3_gx(x):
34 if x[:].sum() < 0:
35 print(x[1:].sum(), x[1:])
36 ans = 1 + 9 / 29 * x[1:].sum()
37 return ans
38
39
40 def zdt3_f2(x):
41 g = zdt3_gx(x)
42 ans = 1 - np.sqrt(x[0] / g) - (x[0] / g) * np.sin(10 * np.pi * x[0])
43 return ans
44
45
46 class Objective_Function:
47 function_dic = defaultdict(lambda: None)
48
49 def __init__(self, f_funcs, domain):
50 self.f_funcs = f_funcs
51 self.domain = domain
52
53 @staticmethod
54 def get_one_function(name):
55 if Objective_Function.function_dic[name] is not None:
56 return Objective_Function.function_dic[name]
57
58 if name == "zdt4":
59 f_funcs = [zdt4_f1, zdt4_f2]
60 domain = [[0, 1]]
61 for i in range(9):
62 domain.append([-5, 5])
63 Objective_Function.function_dic[name] = Objective_Function(f_funcs, domain)
64 return Objective_Function.function_dic[name]
65
66 if name == "zdt3":
67 f_funcs = [zdt3_f1, zdt3_f2]
68 domain = [[0, 1] for i in range(30)]
69 Objective_Function.function_dic[name] = Objective_Function(f_funcs, domain)
70 return Objective_Function.function_dic[name]
Moead_Util.py
1 import numpy as np
2
3 from GA.MOEAD.Person import Person
4
5
6 def distribution_number(sum, m):
7 # 取m个数,数的和为N
8 if m == 1:
9 return [[sum]]
10 vectors = []
11 for i in range(1, sum - (m - 1) + 1):
12 right_vec = distribution_number(sum - i, m - 1)
13 a = [i]
14 for item in right_vec:
15 vectors.append(a + item)
16 return vectors
17
18
19 class Moead_Util:
20 def __init__(self, N, m, T, o_func, pm):
21 self.N = N
22 self.m = m
23 self.T = T # 邻居大小限制
24 self.o_func = o_func
25 self.pm = pm # 变异概率
26
27 self.Z = np.zeros(shape=m)
28
29 self.EP = [] # 前沿
30 self.EP_fx = [] # ep对应的目标值
31 self.weight_vectors = None # 均匀权重向量
32 self.Euler_distance = None # 欧拉距离矩阵
33 self.pip_size = -1
34
35 self.pop = None
36 # self.pop_dna = None
37
38 def init_mean_vector(self):
39 vectors = distribution_number(self.N + self.m, self.m)
40 vectors = (np.array(vectors) - 1) / self.N
41 self.weight_vectors = vectors
42 self.pip_size = len(vectors)
43 return vectors
44
45 def init_Euler_distance(self):
46 vectors = self.weight_vectors
47 v_len = len(vectors)
48
49 Euler_distance = np.zeros((v_len, v_len))
50 for i in range(v_len):
51 for j in range(v_len):
52 distance = ((vectors[i] - vectors[j]) ** 2).sum()
53 Euler_distance[i][j] = distance
54
55 self.Euler_distance = Euler_distance
56 return Euler_distance
57
58 def init_population(self):
59 pop_size = self.pip_size
60 dna_len = len(self.o_func.domain)
61 pop = []
62 pop_dna = np.random.random(size=(pop_size, dna_len))
63 # 初始个体的 dna
64 for i in range(pop_size):
65 pop.append(Person(pop_dna[i]))
66
67 # 初始个体的 weight_vector, neighbor, o_func
68 for i in range(pop_size):
69 # weight_vector, neighbor, o_func
70 person = pop[i]
71 distance = self.Euler_distance[i]
72 sort_arg = np.argsort(distance)
73 weight_vector = self.weight_vectors[i]
74 # neighbor = pop[sort_arg][:self.T]
75 neighbor = []
76 for i in range(self.T):
77 neighbor.append(pop[sort_arg[i]])
78
79 o_func = self.o_func
80 person.set_info(weight_vector, neighbor, o_func)
81 self.pop = pop
82 # self.pop_dna = pop_dna
83
84 return pop
85
86 def init_Z(self):
87 Z = np.full(shape=self.m, fill_value=float("inf"))
88 for person in self.pop:
89 for i in range(len(self.o_func.f_funcs)):
90 f = self.o_func.f_funcs[i]
91 # f_x_i:某个体,在第i目标上的值
92 f_x_i = f(person.dna)
93 if f_x_i < Z[i]:
94 Z[i] = f_x_i
95
96 self.Z = Z
97
98 def get_fx(self, dna):
99 fx = []
100 for f in self.o_func.f_funcs:
101 fx.append(f(dna))
102 return fx
103
104 def update_ep(self, new_dna):
105 # 将新解与EP每一个进行比较,删除被新解支配的
106 # 如果新解没有被旧解支配,则保留
107 new_dna_fx = self.get_fx(new_dna)
108 accept_new = True # 是否将新解加入EP
109 # print(f"准备开始循环: EP长度{len(self.EP)}")
110 for i in range(len(self.EP) - 1, -1, -1): # 从后往前遍历
111 old_ep_item = self.EP[i]
112 old_fx = self.EP_fx[i]
113 # old_fx = self.get_fx(old_ep_item)
114 a_b = True # 老支配行
115 b_a = True # 新支配老
116 for j in range(len(self.o_func.f_funcs)):
117 if old_fx[j] < new_dna_fx[j]:
118 b_a = False
119 if old_fx[j] > new_dna_fx[j]:
120 a_b = False
121 # T T : fx相等 直接不改变EP
122 # T F :老支配新 留老,一定不要新,结束循环.
123 # F T :新支配老 留新,一定不要这个老,继续循环
124 # F F : 非支配关系 不操作,循环下一个
125 # TF为什么结束循环,FT为什么继续循环,你们可以琢磨下
126 if a_b:
127 accept_new = False
128 break
129 if not a_b and b_a:
130 if len(self.EP) <= i:
131 print(len(self.EP), i)
132 del self.EP[i]
133 del self.EP_fx[i]
134 continue
135
136 if accept_new:
137 self.EP.append(new_dna)
138 self.EP_fx.append(new_dna_fx)
139 return self.EP, self.EP_fx
140
141 def update_Z(self, new_dna):
142 new_dna_fx = self.get_fx(new_dna)
143 Z = self.Z
144 for i in range(len(self.o_func.f_funcs)):
145 if new_dna_fx[i] < Z[i]:
146 Z[i] = new_dna_fx[i]
147 return Z
实现.py
import random
import numpy as np
from GA.MOEAD.Moead_Util import Moead_Util
from GA.MOEAD.Objective_Function import Objective_Function
from GA.MOEAD.Person import Person
import matplotlib.pyplot as plt
def draw(x, y):
plt.scatter(x, y, s=10, c="grey") # s 点的大小 c 点的颜色 alpha 透明度
plt.show()
iterations = 1000 # 迭代次数
N = 400
m = 2
T = 5
o_func = Objective_Function.get_one_function("zdt4")
pm = 0.7
moead = Moead_Util(N, m, T, o_func, pm)
moead.init_mean_vector()
moead.init_Euler_distance()
pop = moead.init_population()
moead.init_Z()
for i in range(iterations):
print(i, len(moead.EP))
for person in pop:
p1, p2 = person.choice_two_person()
d1, d2 = Person.cross_get_two_new_dna(p1, p2)
if np.random.rand() < pm:
p1.mutation_dna(d1)
if np.random.rand() < pm:
p1.mutation_dna(d2)
moead.update_Z(d1)
moead.update_Z(d2)
t1, t2 = person.choice_two_person()
if t1.compare(d1) < 0:
t1.accept_new_dna(d1)
moead.update_ep(d1)
if t2.compare(d1) < 0:
t2.accept_new_dna(d2)
moead.update_ep(d1)
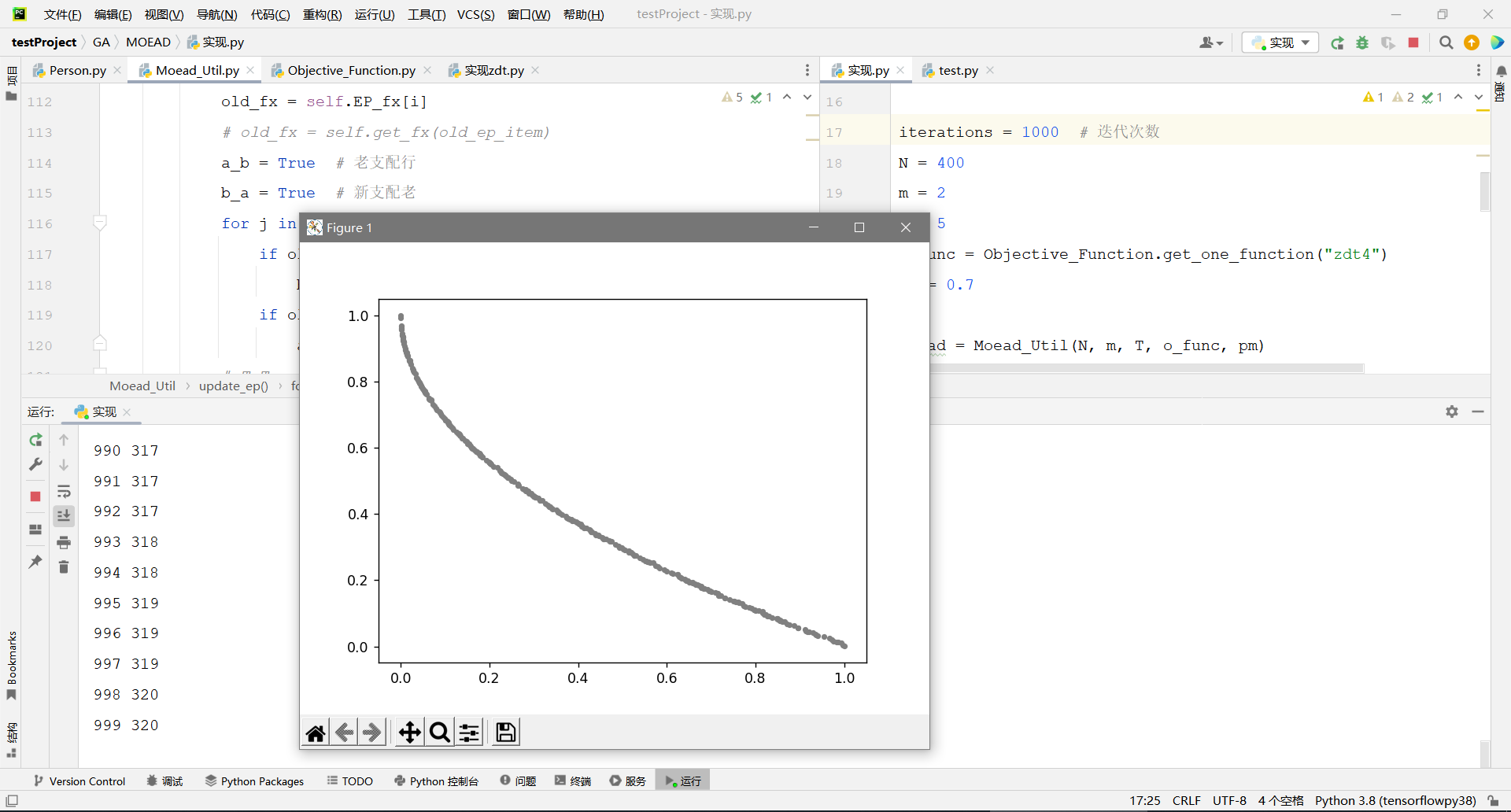
# 输出结果画图
EP_fx = np.array(moead.EP_fx)
x = EP_fx[:, 0]
y = EP_fx[:, 1]
draw(x, y)
效果-ZDT4
本文原创作者:湘潭大学-魏雄,未经许可禁止转载
Recommend
About Joyk
Aggregate valuable and interesting links.
Joyk means Joy of geeK